《计算机应用》唯一官方网站 ›› 2025, Vol. 45 ›› Issue (2): 546-555.DOI: 10.11772/j.issn.1001-9081.2024020177
• 先进计算 • 上一篇
谢冬梅1, 边昕烨1, 于连飞1, 刘文博1, 王子灵1, 曲志坚1( ), 于家峰2
), 于家峰2
收稿日期:2024-02-26
修回日期:2024-04-14
接受日期:2024-04-16
发布日期:2024-06-04
出版日期:2025-02-10
通讯作者:
曲志坚
作者简介:谢冬梅(1998—),女,山东淄博人,硕士研究生,CCF会员,主要研究方向:深度学习、生物信息学基金资助:
Dongmei XIE1, Xinye BIAN1, Lianfei YU1, Wenbo LIU1, Ziling WANG1, Zhijian QU1( ), Jiafeng YU2
), Jiafeng YU2
Received:2024-02-26
Revised:2024-04-14
Accepted:2024-04-16
Online:2024-06-04
Published:2025-02-10
Contact:
Zhijian QU
About author:XIE Dongmei, born in 1998, M. S. candidate. Her research interests include deep learning, bioinformatics.Supported by:摘要:
小开放阅读框(sORFs)在多种生物学过程中发挥着关键作用,且准确识别编码sORFs和非编码sORFs是基因组学中一项重要且有挑战性的任务。针对目前大多数编码sORFs预测算法严重依赖基于先验生物知识的手工特征且缺乏通用性的问题以及原始sORFs的序列长度长短不一而无法直接输入预测模型的问题,提出一种基于sORF-Graph图编码方式的端到端的深度学习框架DeepsORF预测编码sORFs。首先,通过sORF-Graph将所有sORFs序列编码成对应的图,并将序列信息编码成图元素特征,从而对输入序列进行标准化处理;其次,引入基于卷积与残差的流注意力机制捕获sORFs中碱基远距离之间的相互作用,以更有效地表达sORFs的特征,并提高模型的预测精度。实验结果证明,DeepsORF框架在6个独立测试集上的性能均得到提升,与csORF-finder方法相比,DeepsORF在D.melanogaster nonCDS-sORFs测试集上的准确率、马修斯相关系数(MCC)以及精确率分别提升了9.97、19.49与13.07个百分点,验证了DeepsORF模型在识别编码sORFs和非编码sORFs任务中的有效性以及良好泛化能力。
中图分类号:
谢冬梅, 边昕烨, 于连飞, 刘文博, 王子灵, 曲志坚, 于家峰. 基于图编码与改进流注意力的编码sORFs预测方法DeepsORF[J]. 计算机应用, 2025, 45(2): 546-555.
Dongmei XIE, Xinye BIAN, Lianfei YU, Wenbo LIU, Ziling WANG, Zhijian QU, Jiafeng YU. DeepsORF: coding sORFs prediction method based on graph coding with improved flow attention[J]. Journal of Computer Applications, 2025, 45(2): 546-555.
| 数据集 | 注释 | 编码 sORFs数 | 非编码sORFs数 |
|---|---|---|---|
| H.sapiens CDS-sORFs | 训练集 | 7 232 | 7 232 |
| 测试集 | 1 808 | 1 808 | |
| H.sapiens nonCDS-sORFs | 训练集 | 7 750 | 7 750 |
| 测试集 | 1 937 | 1 937 | |
| M.musculus CDS-sORFs | 训练集 | 2 615 | 2 615 |
| 测试集 | 654 | 654 | |
| M.musculus nonCDS-sORFs | 训练集 | 3 066 | 3 066 |
| 测试集 | 767 | 767 | |
| D.melanogaster CDS-sORFs | 训练集 | 682 | 682 |
| 测试集 | 171 | 171 | |
| D.melanogaster nonCDS-sORFs | 训练集 | 6 978 | 6 978 |
| 测试集 | 1 745 | 1 745 |
表1 数据集信息
Tab. 1 Dataset information
| 数据集 | 注释 | 编码 sORFs数 | 非编码sORFs数 |
|---|---|---|---|
| H.sapiens CDS-sORFs | 训练集 | 7 232 | 7 232 |
| 测试集 | 1 808 | 1 808 | |
| H.sapiens nonCDS-sORFs | 训练集 | 7 750 | 7 750 |
| 测试集 | 1 937 | 1 937 | |
| M.musculus CDS-sORFs | 训练集 | 2 615 | 2 615 |
| 测试集 | 654 | 654 | |
| M.musculus nonCDS-sORFs | 训练集 | 3 066 | 3 066 |
| 测试集 | 767 | 767 | |
| D.melanogaster CDS-sORFs | 训练集 | 682 | 682 |
| 测试集 | 171 | 171 | |
| D.melanogaster nonCDS-sORFs | 训练集 | 6 978 | 6 978 |
| 测试集 | 1 745 | 1 745 |
| 数据集 | 指标 | 方法 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CPPred | MiPepid | RNAsamba | DeepCPP | PsORFs | csORF-finder | codingCapacity | ABLNCPP | DeepsORF | ||
H.sapiens CDS-sORFs | SN | 0.612 3 | 0.954 6 | 0.682 5 | 0.588 5 | 0.782 6 | 0.846 2 | 0.787 6 | 0.815 8 | 0.857 9 |
| SP | 0.675 3 | 0.068 0 | 0.709 1 | 0.679 2 | 0.827 4 | 0.820 2 | 0.829 6 | 0.822 5 | 0.870 0 | |
| ACC | 0.643 8 | 0.511 3 | 0.695 8 | 0.633 8 | 0.805 0 | 0.833 2 | 0.806 6 | 0.819 1 | 0.863 9 | |
| MCC | 0.288 2 | 0.049 0 | 0.391 7 | 0.268 8 | 0.610 7 | 0.666 7 | 0.617 8 | 0.638 3 | 0.727 9 | |
| Precision | 0.653 5 | 0.506 0 | 0.701 1 | 0.647 2 | 0.819 3 | 0.824 8 | 0.822 2 | 0.821 3 | 0.868 4 | |
H.sapiens nonCDS-sORFs | SN | 0.594 7 | 0.642 2 | 0.680 9 | 0.490 4 | 0.759 4 | 0.842 5 | 0.767 7 | 0.788 1 | 0.862 7 |
| SP | 0.733 6 | 0.698 5 | 0.805 9 | 0.820 9 | 0.817 2 | 0.811 0 | 0.850 3 | 0.866 3 | 0.916 4 | |
| ACC | 0.664 2 | 0.670 4 | 0.743 4 | 0.655 7 | 0.788 3 | 0.826 8 | 0.809 0 | 0.827 2 | 0.889 5 | |
| MCC | 0.331 6 | 0.341 3 | 0.490 7 | 0.329 8 | 0.577 6 | 0.653 9 | 0.620 1 | 0.656 4 | 0.780 2 | |
| Precision | 0.690 6 | 0.680 5 | 0.778 2 | 0.732 5 | 0.806 0 | 0.816 8 | 0.836 8 | 0.854 8 | 0.911 6 | |
M.musculus CDS-sORFs | SN | 0.712 5 | 0.919 0 | 0.639 1 | 0.587 2 | 0.874 6 | 0.922 0 | 0.859 3 | 0.761 5 | 0.889 9 |
| SP | 0.675 8 | 0.097 9 | 0.776 8 | 0.691 1 | 0.839 4 | 0.816 5 | 0.830 3 | 0.692 7 | 0.879 2 | |
| ACC | 0.694 2 | 0.508 4 | 0.708 0 | 0.639 1 | 0.857 0 | 0.869 3 | 0.844 8 | 0.727 0 | 0.884 6 | |
| MCC | 0.388 6 | 0.029 5 | 0.419 9 | 0.279 8 | 0.714 5 | 0.742 7 | 0.689 9 | 0.455 2 | 0.769 2 | |
| Precision | 0.687 3 | 0.504 6 | 0.741 1 | 0.655 3 | 0.844 9 | 0.834 0 | 0.835 1 | 0.836 1 | 0.880 5 | |
M.musculus nonCDS-sORFs | SN | 0.719 7 | 0.721 0 | 0.760 1 | 0.653 2 | 0.773 1 | 0.860 5 | 0.809 6 | 0.818 3 | 0.890 5 |
| SP | 0.702 7 | 0.585 4 | 0.796 6 | 0.794 0 | 0.857 9 | 0.854 0 | 0.873 5 | 0.901 7 | 0.945 2 | |
| ACC | 0.711 2 | 0.653 2 | 0.778 4 | 0.723 6 | 0.815 5 | 0.857 2 | 0.841 6 | 0.859 9 | 0.917 9 | |
| MCC | 0.422 5 | 0.309 2 | 0.557 1 | 0.451 7 | 0.633 3 | 0.714 5 | 0.684 6 | 0.722 5 | 0.837 0 | |
| Precision | 0.707 7 | 0.634 9 | 0.788 9 | 0.760 2 | 0.844 7 | 0.854 9 | 0.864 9 | 0.893 0 | 0.942 1 | |
D.melanogaster CDS-sORFs | SN | 0.643 3 | 0.970 8 | 0.707 6 | 0.584 8 | 0.842 1 | 0.812 9 | 0.842 1 | 0.733 7 | 0.853 8 |
| SP | 0.725 1 | 0.017 5 | 0.701 8 | 0.672 5 | 0.888 9 | 0.742 7 | 0.871 3 | 0.736 5 | 0.883 0 | |
| ACC | 0.684 2 | 0.494 2 | 0.704 7 | 0.628 7 | 0.865 5 | 0.777 8 | 0.856 7 | 0.735 1 | 0.868 4 | |
| MCC | 0.369 7 | -0.038 7 | 0.409 4 | 0.258 3 | 0.731 8 | 0.556 9 | 0.713 8 | 0.470 2 | 0.737 2 | |
| Precision | 0.700 6 | 0.497 0 | 0.703 5 | 0.641 0 | 0.883 4 | 0.759 6 | 0.867 5 | 0.738 1 | 0.879 5 | |
D.melanogaster nonCDS-sORFs | SN | 0.111 2 | 0.506 | 0.598 3 | 0.180 5 | 0.695 1 | 0.803 4 | 0.710 6 | 0.781 7 | 0.830 4 |
| SP | 0.876 2 | 0.622 3 | 0.522 1 | 0.780 5 | 0.703 2 | 0.665 3 | 0.746 1 | 0.726 9 | 0.837 8 | |
| ACC | 0.493 7 | 0.564 2 | 0.560 2 | 0.480 5 | 0.699 1 | 0.734 4 | 0.728 4 | 0.754 3 | 0.834 1 | |
| MCC | -0.019 6 | 0.129 2 | 0.120 7 | -0.048 7 | 0.398 3 | 0.473 3 | 0.457 0 | 0.509 3 | 0.668 2 | |
| Precision | 0.473 2 | 0.572 6 | 0.555 9 | 0.451 3 | 0.700 8 | 0.705 9 | 0.736 8 | 0.741 3 | 0.836 6 | |
表2 DeepsORF与对比方法在6个独立测试集上的性能比较
Tab. 2 Performance comparison between DeepsORF and comparative methods on six independent test sets
| 数据集 | 指标 | 方法 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CPPred | MiPepid | RNAsamba | DeepCPP | PsORFs | csORF-finder | codingCapacity | ABLNCPP | DeepsORF | ||
H.sapiens CDS-sORFs | SN | 0.612 3 | 0.954 6 | 0.682 5 | 0.588 5 | 0.782 6 | 0.846 2 | 0.787 6 | 0.815 8 | 0.857 9 |
| SP | 0.675 3 | 0.068 0 | 0.709 1 | 0.679 2 | 0.827 4 | 0.820 2 | 0.829 6 | 0.822 5 | 0.870 0 | |
| ACC | 0.643 8 | 0.511 3 | 0.695 8 | 0.633 8 | 0.805 0 | 0.833 2 | 0.806 6 | 0.819 1 | 0.863 9 | |
| MCC | 0.288 2 | 0.049 0 | 0.391 7 | 0.268 8 | 0.610 7 | 0.666 7 | 0.617 8 | 0.638 3 | 0.727 9 | |
| Precision | 0.653 5 | 0.506 0 | 0.701 1 | 0.647 2 | 0.819 3 | 0.824 8 | 0.822 2 | 0.821 3 | 0.868 4 | |
H.sapiens nonCDS-sORFs | SN | 0.594 7 | 0.642 2 | 0.680 9 | 0.490 4 | 0.759 4 | 0.842 5 | 0.767 7 | 0.788 1 | 0.862 7 |
| SP | 0.733 6 | 0.698 5 | 0.805 9 | 0.820 9 | 0.817 2 | 0.811 0 | 0.850 3 | 0.866 3 | 0.916 4 | |
| ACC | 0.664 2 | 0.670 4 | 0.743 4 | 0.655 7 | 0.788 3 | 0.826 8 | 0.809 0 | 0.827 2 | 0.889 5 | |
| MCC | 0.331 6 | 0.341 3 | 0.490 7 | 0.329 8 | 0.577 6 | 0.653 9 | 0.620 1 | 0.656 4 | 0.780 2 | |
| Precision | 0.690 6 | 0.680 5 | 0.778 2 | 0.732 5 | 0.806 0 | 0.816 8 | 0.836 8 | 0.854 8 | 0.911 6 | |
M.musculus CDS-sORFs | SN | 0.712 5 | 0.919 0 | 0.639 1 | 0.587 2 | 0.874 6 | 0.922 0 | 0.859 3 | 0.761 5 | 0.889 9 |
| SP | 0.675 8 | 0.097 9 | 0.776 8 | 0.691 1 | 0.839 4 | 0.816 5 | 0.830 3 | 0.692 7 | 0.879 2 | |
| ACC | 0.694 2 | 0.508 4 | 0.708 0 | 0.639 1 | 0.857 0 | 0.869 3 | 0.844 8 | 0.727 0 | 0.884 6 | |
| MCC | 0.388 6 | 0.029 5 | 0.419 9 | 0.279 8 | 0.714 5 | 0.742 7 | 0.689 9 | 0.455 2 | 0.769 2 | |
| Precision | 0.687 3 | 0.504 6 | 0.741 1 | 0.655 3 | 0.844 9 | 0.834 0 | 0.835 1 | 0.836 1 | 0.880 5 | |
M.musculus nonCDS-sORFs | SN | 0.719 7 | 0.721 0 | 0.760 1 | 0.653 2 | 0.773 1 | 0.860 5 | 0.809 6 | 0.818 3 | 0.890 5 |
| SP | 0.702 7 | 0.585 4 | 0.796 6 | 0.794 0 | 0.857 9 | 0.854 0 | 0.873 5 | 0.901 7 | 0.945 2 | |
| ACC | 0.711 2 | 0.653 2 | 0.778 4 | 0.723 6 | 0.815 5 | 0.857 2 | 0.841 6 | 0.859 9 | 0.917 9 | |
| MCC | 0.422 5 | 0.309 2 | 0.557 1 | 0.451 7 | 0.633 3 | 0.714 5 | 0.684 6 | 0.722 5 | 0.837 0 | |
| Precision | 0.707 7 | 0.634 9 | 0.788 9 | 0.760 2 | 0.844 7 | 0.854 9 | 0.864 9 | 0.893 0 | 0.942 1 | |
D.melanogaster CDS-sORFs | SN | 0.643 3 | 0.970 8 | 0.707 6 | 0.584 8 | 0.842 1 | 0.812 9 | 0.842 1 | 0.733 7 | 0.853 8 |
| SP | 0.725 1 | 0.017 5 | 0.701 8 | 0.672 5 | 0.888 9 | 0.742 7 | 0.871 3 | 0.736 5 | 0.883 0 | |
| ACC | 0.684 2 | 0.494 2 | 0.704 7 | 0.628 7 | 0.865 5 | 0.777 8 | 0.856 7 | 0.735 1 | 0.868 4 | |
| MCC | 0.369 7 | -0.038 7 | 0.409 4 | 0.258 3 | 0.731 8 | 0.556 9 | 0.713 8 | 0.470 2 | 0.737 2 | |
| Precision | 0.700 6 | 0.497 0 | 0.703 5 | 0.641 0 | 0.883 4 | 0.759 6 | 0.867 5 | 0.738 1 | 0.879 5 | |
D.melanogaster nonCDS-sORFs | SN | 0.111 2 | 0.506 | 0.598 3 | 0.180 5 | 0.695 1 | 0.803 4 | 0.710 6 | 0.781 7 | 0.830 4 |
| SP | 0.876 2 | 0.622 3 | 0.522 1 | 0.780 5 | 0.703 2 | 0.665 3 | 0.746 1 | 0.726 9 | 0.837 8 | |
| ACC | 0.493 7 | 0.564 2 | 0.560 2 | 0.480 5 | 0.699 1 | 0.734 4 | 0.728 4 | 0.754 3 | 0.834 1 | |
| MCC | -0.019 6 | 0.129 2 | 0.120 7 | -0.048 7 | 0.398 3 | 0.473 3 | 0.457 0 | 0.509 3 | 0.668 2 | |
| Precision | 0.473 2 | 0.572 6 | 0.555 9 | 0.451 3 | 0.700 8 | 0.705 9 | 0.736 8 | 0.741 3 | 0.836 6 | |
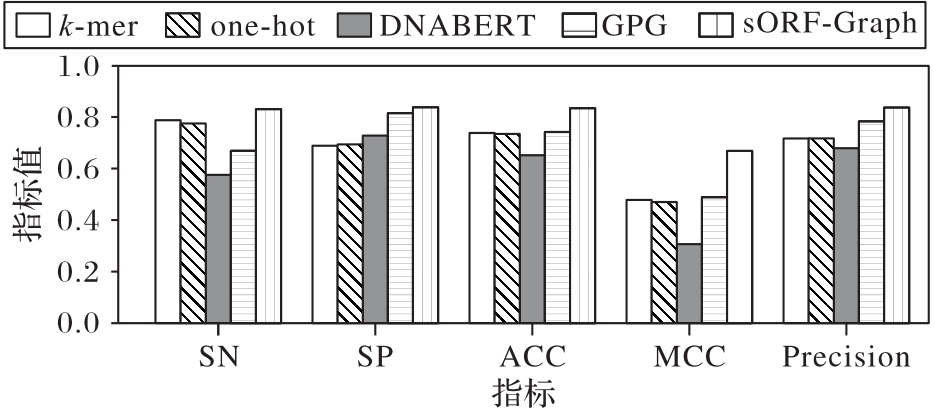
图8 不同编码方式在D.melanogaster nonCDS-sORFs数据集上的性能指标比较
Fig. 8 Performance indicator comparison of different encoding methods on D.melanogaster nonCDS-sORFs dataset
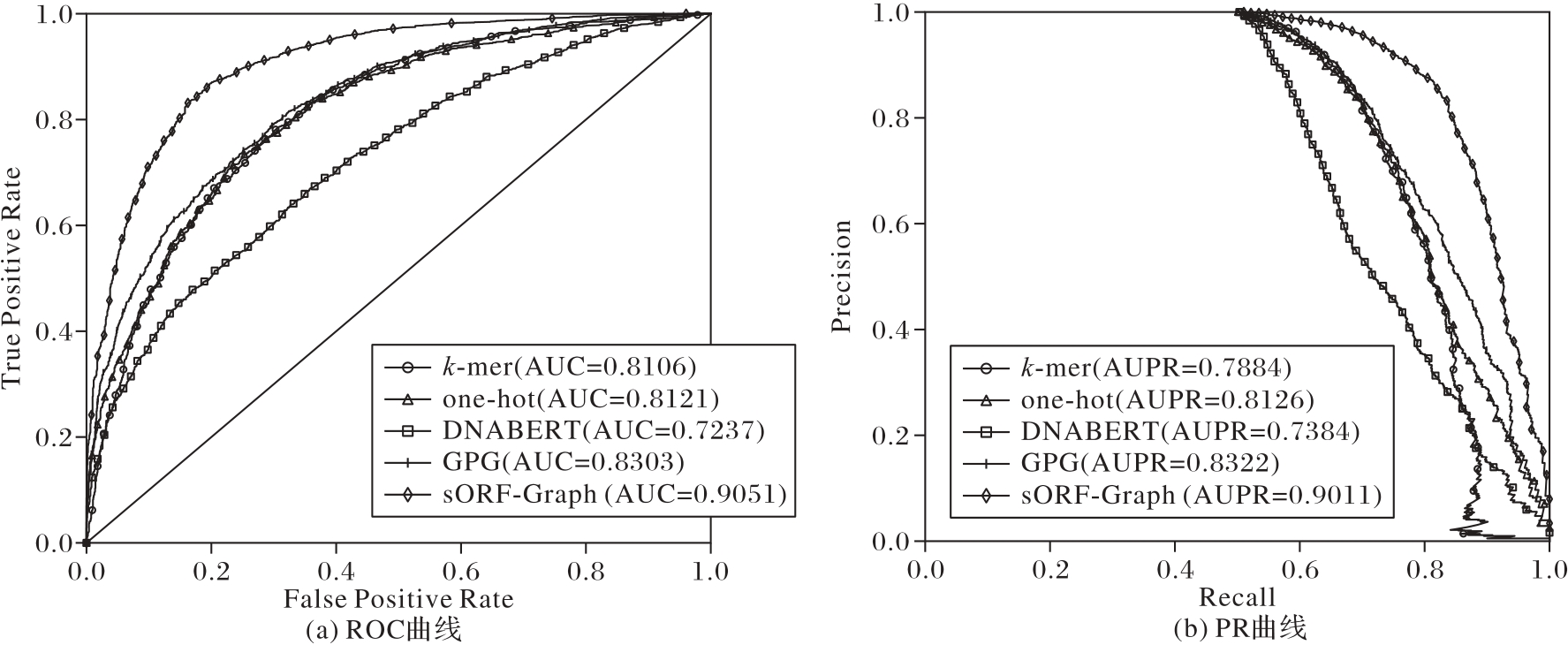
图9 不同编码方式在D.melanogaster nonCDS-sORFs数据集上的ROC和PR曲线比较
Fig. 9 ROC and PR curve comparison of different encoding methods on D.melanogaster nonCDS-sORFs dataset
| 数据集 | 方法 | SN | SP | ACC | MCC | 精确率 |
|---|---|---|---|---|---|---|
| H.sapiens CDS-sORFs | Flow attention | 0.844 6 | 0.850 7 | 0.847 6 | 0.695 3 | 0.849 7 |
| CR-Flow attention | 0.857 9 | 0.870 0 | 0.863 9 | 0.727 9 | 0.868 4 | |
| H.sapiens nonCDS-sORFs | Flow attention | 0.850 8 | 0.921 0 | 0.885 9 | 0.773 7 | 0.915 0 |
| CR-Flow attention | 0.862 7 | 0.916 4 | 0.889 5 | 0.780 2 | 0.911 6 | |
| M.musculus CDS-sORFs | Flow attention | 0.854 7 | 0.880 7 | 0.867 7 | 0.735 7 | 0.877 6 |
| CR-Flow attention | 0.889 9 | 0.879 2 | 0.884 6 | 0.769 2 | 0.880 5 | |
| M.musculus nonCDS-sORFs | Flow attention | 0.900 9 | 0.936 1 | 0.918 5 | 0.837 5 | 0.933 8 |
| CR-Flow attention | 0.890 5 | 0.945 2 | 0.917 9 | 0.837 0 | 0.942 1 | |
| D.melanogaster CDS-sORFs | Flow attention | 0.836 3 | 0.777 8 | 0.807 0 | 0.615 1 | 0.790 1 |
| CR-Flow attention | 0.853 8 | 0.883 0 | 0.868 4 | 0.737 2 | 0.879 5 | |
| D.melanogaster nonCDS-sORFs | Flow attention | 0.813 2 | 0.825 2 | 0.819 2 | 0.638 4 | 0.823 1 |
| CR-Flow attention | 0.830 4 | 0.837 8 | 0.834 1 | 0.668 2 | 0.836 6 |
表3 流注意力与基于卷积与残差的流注意力性能比较
Tab. 3 Performance comparison of Flow attention and CR-Flow attention
| 数据集 | 方法 | SN | SP | ACC | MCC | 精确率 |
|---|---|---|---|---|---|---|
| H.sapiens CDS-sORFs | Flow attention | 0.844 6 | 0.850 7 | 0.847 6 | 0.695 3 | 0.849 7 |
| CR-Flow attention | 0.857 9 | 0.870 0 | 0.863 9 | 0.727 9 | 0.868 4 | |
| H.sapiens nonCDS-sORFs | Flow attention | 0.850 8 | 0.921 0 | 0.885 9 | 0.773 7 | 0.915 0 |
| CR-Flow attention | 0.862 7 | 0.916 4 | 0.889 5 | 0.780 2 | 0.911 6 | |
| M.musculus CDS-sORFs | Flow attention | 0.854 7 | 0.880 7 | 0.867 7 | 0.735 7 | 0.877 6 |
| CR-Flow attention | 0.889 9 | 0.879 2 | 0.884 6 | 0.769 2 | 0.880 5 | |
| M.musculus nonCDS-sORFs | Flow attention | 0.900 9 | 0.936 1 | 0.918 5 | 0.837 5 | 0.933 8 |
| CR-Flow attention | 0.890 5 | 0.945 2 | 0.917 9 | 0.837 0 | 0.942 1 | |
| D.melanogaster CDS-sORFs | Flow attention | 0.836 3 | 0.777 8 | 0.807 0 | 0.615 1 | 0.790 1 |
| CR-Flow attention | 0.853 8 | 0.883 0 | 0.868 4 | 0.737 2 | 0.879 5 | |
| D.melanogaster nonCDS-sORFs | Flow attention | 0.813 2 | 0.825 2 | 0.819 2 | 0.638 4 | 0.823 1 |
| CR-Flow attention | 0.830 4 | 0.837 8 | 0.834 1 | 0.668 2 | 0.836 6 |
| 1 | SIEBER P, PLATZER M, SCHUSTER S. The definition of open reading frame revisited[J]. Trends in Genetics, 2018, 34(3): 167-170. |
| 2 | BASRAI M A, HIETER P, BOEKE J F. Small open reading frames: beautiful needles in the haystack[J]. Genome Research, 1997, 7(8): 768-771. |
| 3 | ORR M W, MAO Y, STORZ G, et al. Alternative ORFs and small ORFs: shedding light on the dark proteome[J]. Nucleic Acids Res, 2020, 48(3): 1029-1042. |
| 4 | GALINDO M I, PUEYO J I, FOUIX S, et al. Peptides encoded by short ORFs control development and define a new eukaryotic gene family[J]. PLoS Biology, 2007, 5(5): No.e106. |
| 5 | COUSO J P, PATRAQUIM P. Classification and function of small open reading frames[J]. Nature Reviews Molecular Cell Biology, 2017, 18(9): 575-589. |
| 6 | HANADA K, AKIYAMA K, SAKURAI T, et al. sORF finder: a program package to identify small open reading frames with high coding potential[J]. Bioinformatics, 2010, 26(3): 399-400. |
| 7 | TONG X, LIU S. CPPred: coding potential prediction based on the global description of RNA sequence[J]. Nucleic Acids Research, 2019, 47(8): No.e43. |
| 8 | ZHU M, GRIBSKOV M. MiPepid: MicroPeptide identification tool using machine learning[J]. BMC Bioinformatics, 2019, 20: No.559. |
| 9 | TONG X, HONG X, XIE J, et al. CPPred-sORF: coding potential prediction of sORF based on non-AUG[EB/OL]. [2024-05-23].. |
| 10 | YU J, GUO L, DOU X, et al. Comprehensive evaluation of protein-coding sORFs prediction based on a random sequence strategy[J]. Frontiers in Bioscience-Landmark, 2021, 26(8): 272-278. |
| 11 | YU J, JIANG W, ZHU S B, et al. Prediction of protein-coding small ORFs in multi-species using integrated sequence-derived features and the random forest model[J]. Methods, 2023, 210: 10-19. |
| 12 | ZHAO S, MENG J, WEKESA J S, et al. Identification of small open reading frames in plant lncRNA using class-imbalance learning[J]. Computers in Biology and Medicine, 2023, 157: No.106773. |
| 13 | ZHANG Y, JIA C, FULLWOOD M J, et al. DeepCPP: a deep neural network based on nucleotide bias information and minimum distribution similarity feature selection for RNA coding potential prediction[J]. Briefings in Bioinformatics, 2021, 22(2): 2073-2084. |
| 14 | CAMARGO A P, SOURKOV V, PEREIRA G A G, et al. RNAsamba: neural network-based assessment of the protein-coding potential of RNA sequences[J]. NAR Genomics and Bioinformatics, 2020, 2(1): No.lqz024. |
| 15 | ZHANG M, ZHAO J, LI C, et al. csORF-finder: an effective ensemble learning framework for accurate identification of multi-species coding short open reading frames[J]. Briefings in Bioinformatics, 2022, 23(6): No.bbac392. |
| 16 | DENG L, JIANG Y, HU X, et al. ABLNCPP: attention mechanism-based bidirectional long short-term memory for noncoding RNA coding potential prediction[J]. Journal of Chemical Information and Modeling, 2023, 63(12): 3955-3966. |
| 17 | KHANDUJA A, KUMAR M, MOHANTY D. ProsmORF-pred: a machine learning-based method for the identification of small ORFs in prokaryotic genomes[J]. Briefings in Bioinformatics, 2023, 24(3): No.bbad101. |
| 18 | WANG X, GAO X, WANG G, et al. miProBERT: identification of microRNA promoters based on the pre-trained model BERT[J]. Briefings in Bioinformatics, 2023, 24(3): No.bbad093. |
| 19 | LIU X, SONG C, HUANG F, et al. GraphCDR: a graph neural network method with contrastive learning for cancer drug response prediction[J]. Briefings in Bioinformatics, 2021, 23(1): No.bbab457. |
| 20 | MA A, WANG X, LI J, et al. Single-cell biological network inference using a heterogeneous graph transformer[J]. Nature Communications, 2023, 14: No.964. |
| 21 | WU Y, GAO M, ZENG M, et al. BridgeDPI: a novel graph neural network for predicting drug-protein interactions[J]. Bioinformatics, 2022, 38(9): 2571-2578. |
| 22 | 华阳,李金星,冯振华,等. 注意力特征融合的蛋白质-药物相互作用预测[J]. 计算机研究与发展, 2022, 59(9): 2051-2065. |
| HUA Y, LI J X, FENG Z H, et al. Protein-drug interaction prediction based on attention feature fusion[J]. Journal of Computer Research and Development, 2022, 59(9): 2051-2065. | |
| 23 | 陶斯涵,丁彦蕊. 引入序列信息的残基相互作用网络比对算法[J]. 软件学报, 2019, 30(11): 3413-3426. |
| TAO S H, DING Y R. Algorithm introduced sequence information for residue interaction network alignment[J]. Journal of Software, 2019, 30(11): 3413-3426. | |
| 24 | WANG R, NG Y K, ZHANG X, et al. A graph representation of gapped patterns in phage sequences for graph convolutional network[EB/OL]. [2024-05-23].. |
| 25 | LI A, ZHANG J, ZHOU Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme[J]. BMC Bioinformatics, 2014, 15: No.311. |
| 26 | DEVLIN J, CHANG M W, LEE K, et al. BERT: pre-training of deep bidirectional transformers for language understanding[C]// Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). Stroudsburg: ACL, 2019: 4171-4186. |
| 27 | JI Y, ZHOU Z, LIU H, et al. DNABERT: pre-trained bidirectional encoder representations from transformers model for DNA-language in genome[J]. Bioinformatics, 2021, 37(15): 2112-2120. |
| 28 | CHANG L, MA C, SUN K, et al. Enhanced road information representation in graph recurrent network for traffic speed prediction[J]. IET Intelligent Transport Systems, 2023, 17(7): 1434-1453. |
| 29 | 牟长宁,王海鹏,周丕宇,等. 基于图卷积神经网络的串联质谱从头测序[J]. 计算机应用, 2021, 41(9): 2773-2779. |
| MOU C N, WANG H P, ZHOU P Y, et al. De novo peptide sequencing by tandem mass spectrometry based on graph convolutional neural network[J]. Journal of Computer Applications, 2021, 41(9): 2773-2779. | |
| 30 | GUO Y, LUO X, CHEN L, et al. DNA-GCN: graph convolutional networks for predicting DNA-protein binding[C]// Proceedings of the 2021 International Conference on Intelligent Computing, LNCS 12838. Cham: Springer, 2021: 458-466. |
| 31 | WU H, WU J, XU J, et al. Flowformer: linearizing transformers with conservation flows[C]// Proceedings of the 39th International Conference on Machine Learning. New York: JMLR.org, 2022: 24226-24242. |
| 32 | YAO Z, ZHANG W, SONG P, et al. DeepFormer: a hybrid network based on convolutional neural network and flow-attention mechanism for identifying the function of DNA sequences[J]. Briefings in Bioinformatics, 2023, 24(2): No.bbad095. |
| 33 | GUNEL B, DU J, CONNEAU A, et al. Supervised contrastive learning for pre-trained language model fine-tuning[EB/OL]. [2024-04-02].. |
| [1] | 蒋铭, 王琳钦, 赖华, 高盛祥. 基于编辑约束的端到端越南语文本正则化方法[J]. 《计算机应用》唯一官方网站, 2025, 45(2): 362-370. |
| [2] | 付强, 徐振平, 盛文星, 叶青. 结合字节级别字节对编码的端到端中文语音识别方法[J]. 《计算机应用》唯一官方网站, 2025, 45(1): 318-324. |
| [3] | 赵晓焱, 匡燕, 王梦含, 袁培燕. 基于知识图谱的端到端内容共享机制[J]. 《计算机应用》唯一官方网站, 2024, 44(4): 995-1001. |
| [4] | 尹春勇, 李荧. 基于BCU-Tree与字典的高效用挖掘快速脱敏算法[J]. 《计算机应用》唯一官方网站, 2023, 43(2): 413-422. |
| [5] | 刘聪, 万根顺, 高建清, 付中华. 基于韵律特征辅助的端到端语音识别方法[J]. 《计算机应用》唯一官方网站, 2023, 43(2): 380-384. |
| [6] | 杨磊, 赵红东, 于快快. 基于多头注意力机制的端到端语音情感识别[J]. 《计算机应用》唯一官方网站, 2022, 42(6): 1869-1875. |
| [7] | 郭帅, 苏旸. 基于数据流的加密流量分类方法[J]. 计算机应用, 2021, 41(5): 1386-1391. |
| [8] | 纪腾其, 孟军, 赵思远, 胡鹤还. 基于表示学习和深度森林的长链非编码RNA编码短肽预测模型[J]. 《计算机应用》唯一官方网站, 2021, 41(12): 3614-3619. |
| [9] | 吴赛赛, 梁晓贺, 谢能付, 周爱莲, 郝心宁. 面向领域实体关系联合抽取的标注方法[J]. 计算机应用, 2021, 41(10): 2858-2863. |
| [10] | 胡学敏, 童秀迟, 郭琳, 张若晗, 孔力. 基于深度视觉注意神经网络的端到端自动驾驶模型[J]. 计算机应用, 2020, 40(7): 1926-1931. |
| [11] | 陈修凯, 陆志华, 周宇. 基于卷积编解码器和门控循环单元的语音分离算法[J]. 计算机应用, 2020, 40(7): 2137-2141. |
| [12] | 郝志峰, 柯妍蓉, 李烁, 蔡瑞初, 温雯, 王丽娟. 基于图编码网络的社交网络节点分类方法[J]. 计算机应用, 2020, 40(1): 188-195. |
| [13] | 贾永超, 何小卫, 郑忠龙. 融合重检测机制的卷积回归网络目标跟踪算法[J]. 计算机应用, 2019, 39(8): 2247-2251. |
| [14] | 文凯, 谭笑. 基于用户偏好与副本阈值的端到端缓存算法[J]. 计算机应用, 2019, 39(7): 2051-2055. |
| [15] | 邱泽宇, 屈丹, 张连海. 基于WaveNet的端到端语音合成方法[J]. 计算机应用, 2019, 39(5): 1325-1329. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||